-
PROVIDERS
New MRD Medicare Coverage for Select Indications*
*When coverage criteria are met. Additional criteria and exceptions for coverage may apply.
-
LIFE SCIENCES
REGISTER NOW
UPCOMING WEBINAR
Driving enterprise value with RWD -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
12/01/2023
Q&A: Impact of multimodal data in translational science
Analysis of real-world data (RWD) streamlines translational research and enables providers and researchers to provide precision medicine to patients at scale.
Authors
Pedram Razavi, MD, PhD
Director, Breast Translational Program and Molecular Tumor Board, MSK

Charles Lin, Ph.D.
Senior Vice President, TRN Mapping, KronosBio

Jonathan Dry
Vice President, Pharma R&D, Tempus

Director, Breast Translational Program and Molecular Tumor Board, MSK

Charles Lin, Ph.D.
Senior Vice President, TRN Mapping, KronosBio

Jonathan Dry
Vice President, Pharma R&D, Tempus

Recently, Iker Huerga, EVP of Life Sciences Strategy & RWD at Tempus and Jonathan Dry, Vice President, Pharma R&D at Tempus, sat down with Pedram Razavi, MD, PhD Director, Breast Translational Program and Molecular Tumor Board, MSK and Charles Lin, Ph.D. Senior Vice President, TRN Mapping at KronosBio to discuss the impact and applications of multimodal data in translational science—and how academia and industry are utilizing Tempus data and resources to improve their scientific research efforts.
Iker: Could you please introduce yourself and tell us about what you do? |
| Dr. Razavi: I’m a breast oncologist at MSK, and I lead the Breast Translational Program, which focuses on conducting clinical, genomic, and multimodal analyses to understand the biology of breast cancer, including mechanisms of response and resistance to therapy. Today, I’ll be talking about our experience with the translational program.
Dr. Lin: I’m a computational biologist by training, and I transitioned from academia to industry at KronosBio, a clinical-stage company using small molecules to drug transcription in cancer. It’s been a privilege to work with the team at Tempus across the discovery, translation, and, now, clinical fronts over the last two years. I’m excited to talk about some of that work, especially following the release of our recent clinical data. Jonathan: I’m VP of Pharma R&D at Tempus, and I oversee scientific projects and broader science strategy for our large collaborations with big pharma. Our teams help these companies leverage Tempus’ multimodal real-world data and data generation platforms to accelerate and progress their portfolios by generating actionable insights or producing the right data. |
Iker: So, how are you utilizing multimodal data? |
| Dr. Razavi: Although multimodal data has existed in the oncology space for many years, the program at MSK only started in 2013 with the clinical genomics program, which has since enabled us to sequence early-stage and metastatic breast cancer patients at scale. In 2014, we realized we could match our existing high-quality clinical data with genomic data to better understand the biology of breast cancer.
The feasibility of these studies was validated by proof-of-concept experiments conducted in patients treated with endocrine resistance therapy, where we identified novel mechanisms of resistance and their associated mutations and alterations. By conducting large-scale genomics and transcriptomics analyses, we could study the mechanisms of cyclin-dependent kinase (CDK)4/6 response and resistance to antibody-drug conjugates (ADCs) and other targeted therapies while predicting patient outcomes and prognosis. Dr. Lin: At Kronos Bio, we took the alternative approach, attempting to translate the insight from basic mechanistic studies into clinical development. We are interested in targeting transcription, which is dysregulated in multiple cancers yet essential for cellular function. But without a therapeutic index, it’s challenging to target different transcription factors. At KronosBio, we are targeting CDK9-mediated transcription regulation, which often goes awry in cancers but is difficult to detect using readouts like pathology. Partnering with Tempus helped us leverage multimodal data to find target populations for various indications. We had a CDK9 inhibitor and a mechanistic model that suggested CDK9 dependence in certain cancers but did not have the preclinical models to treat organoids, identify the mutational and transcriptomic changes in pathway properties, and determine the prevalence of transcription deregulation to target clinically. Tempus’ RWD enabled us to hypothesize that transcriptional deregulation mediated by MYC—the most common oncogene and transcription factor—might be sensitive to CDK9 inhibition. A large portion of the patients with “triple negative” ovarian and lung cancers in these multimodal data showed elevated MYC levels, potentially rendering them sensitive to our drug. Using genomic amplification and overexpression, we characterized these tumors and expanded our cohort to these patients. Additionally, Tempus’ TIME clinical trial network helped us build a real-world understanding of where to find verified patients with molecular evidence of rare MYC-deregulated transcription factor fusions that are challenging to detect outside of RNA assays. This work has resulted in our recently released Phase I clinical data, whose safety profile and responses validate our CDK9 attenuation hypothesis. Jonathan: At Tempus, we leverage our understanding of complex biomarker behavior for specific drugs or targets to evaluate shifts following exposure to current standards of care and identify avenues for new agents to act. This can be powerful when designing clinical trials in select indications or therapies. This requires traditional biomarker benchmarking to determine the right populations that might benefit from new agents, and can benefit from organoid-based mechanistic validation to develop representative models of these disease indications and test which combinations of perturbations will work best in these scenarios. It’s also important to have established clinical platforms to increase the accessibility of genomic profiling and enable us to translate the understanding of the association between disease and multimodal biomarkers into clinical practice. More companies are now realizing that multimodal data is crucial to understanding the complexity of tumor biology and strategically targeting cancers via a multidimensional approach. Being able to correlate resistance drivers to the various phenotypes of response observed also helps streamline these studies. |
Iker: What are some practical examples of the role played by multimodal data in improving your translational research? |
| Dr. Lin: While identifying the MYC regulator, we learned that analyzing multimodal data provides a more comprehensive understanding of mechanistic disease biology than using a single modality. Studying the effects of drugging a transcriptional factor like MYC, which regulates hundreds to thousands of genes, requires a multidimensional phenotype readout because the perturbation attempts to selectively alter the tumor’s gene expression program. Understanding how this approach works—and differentiates itself from non-selectively killing cells or pan-transcriptionally inhibiting them—is crucial. With Tempus’ cohort of RNAseq data, we were able to conduct longitudinal analyses of biomarkers and predictively model how we select patients for clinical trials.
As the field continues to evolve from non-specific agents like cytotoxic chemotherapy into more targeted therapies, we can now learn how to predict the potential behavior of cancers and stay a step ahead of it. For example, when looking to validate the findings from studies in which patients were stratified into responders vs. non-responders, multimodal data can provide insights into mechanistic tumor biology, which then helps oncology research teams to systematically probe factors like protein interactions, drug binding kinetics, etc. Dr. Razavi: Using RWD, we’ve learned more about the biology of breast cancer and translated these learnings to improve clinical trial design. For example, we can identify which patient subgroups are most likely to benefit from a therapy versus those that wouldn’t or decide which patients would benefit from escalating or de-escalating medications. Let’s consider patients with estrogen receptor-positive (ER+) breast cancers treated with CDK4/6 inhibitors like Palbociclib and Abemaciclib. Although these medications are transformative therapeutics, they have been clinically administered without any biomarker analysis and with the understanding that 10%-20% of patients didn’t respond well and another 10%-20% barely achieved a median response post-treatment. At MSK, we questioned whether we could identify CDK4/6 inhibitor non-responders, and in our initial analysis, we observed a loss of RB1, which was already biologically validated but also implicated in the Hippo pathway. These findings have since resulted in the development of therapies targeting the Hippo pathway in breast cancer. And we have expanded our clinical program based on the patients who developed CDK4/6 resistance. With thousands of patient data points, we can identify multiple resistance mechanisms, such as PTEN loss. Additionally, multimodal data helps cross-validate findings from different RWD analyses. For instance, Tempus helped validate our finding that BRCA1/2 germline variants result in CDK4/6 resistance, which is now driving the development of PARP inhibitors in this setting. Traditionally, oncologists study whether specific pathways implicated in cancer types are amenable to being targeted, or preclude patients from benefiting from certain therapies—such as KRAS mutant colorectal cancers. Multimodal data is changing the course of oncology care by enabling oncologists to improve their predictions of cancer outcomes and the mechanisms by which these cancers progress across patient subgroups. For instance, we now know that not every ER+ breast cancer patient receiving endocrine therapy will develop estrogen receptor 1 (ESR1) resistance mutations, even after years of treatment; only about 40% of treated breast cancer patients develop them. Understanding these patterns is crucial to designing clinical trials and determining the appropriate dose change or sequence of interventions. |
Iker: How do you think multimodal data will transform the future of translational research? |
| Dr. Lin: It would be great to achieve a robust pipeline through which patients can be enrolled in clinical trials based on similar characterizations of patient and multimodal data models.
Dr. Razavi: I agree. It would be more meaningful for patients if researchers consistently translated findings from multimodal RWD to experimental approaches and vice versa as a tool for stratifying clinical trials, improving existing medications, or identifying new targets. Jonathan: I also agree, but I’d like to add that we’re introducing complexity to the care path for patients and clinicians. However, there’s an opportunity to improve how patients receive care by using multimodal data and AI models to identify the most helpful care path and improve that for future success. |
-
11/11/2025
A new era of biopharma R&D: The TechBio revolution—realities and the next frontier
Join Tempus and Recursion leaders to explore their strategic TechBio partnership. Learn how they use AI and supercomputing with petabytes of data to accelerate drug discovery and development. See the impact on biopharma R&D's future.
Watch replay -
11/14/2025
Validating a novel target and informing trial design for a first-in-class therapeutic
Discover how a biopharma company used Tempus’ de-identified multimodal data to support validation of a novel target and inform trial design for a first-in-class therapeutic.
Read more -
11/14/2025
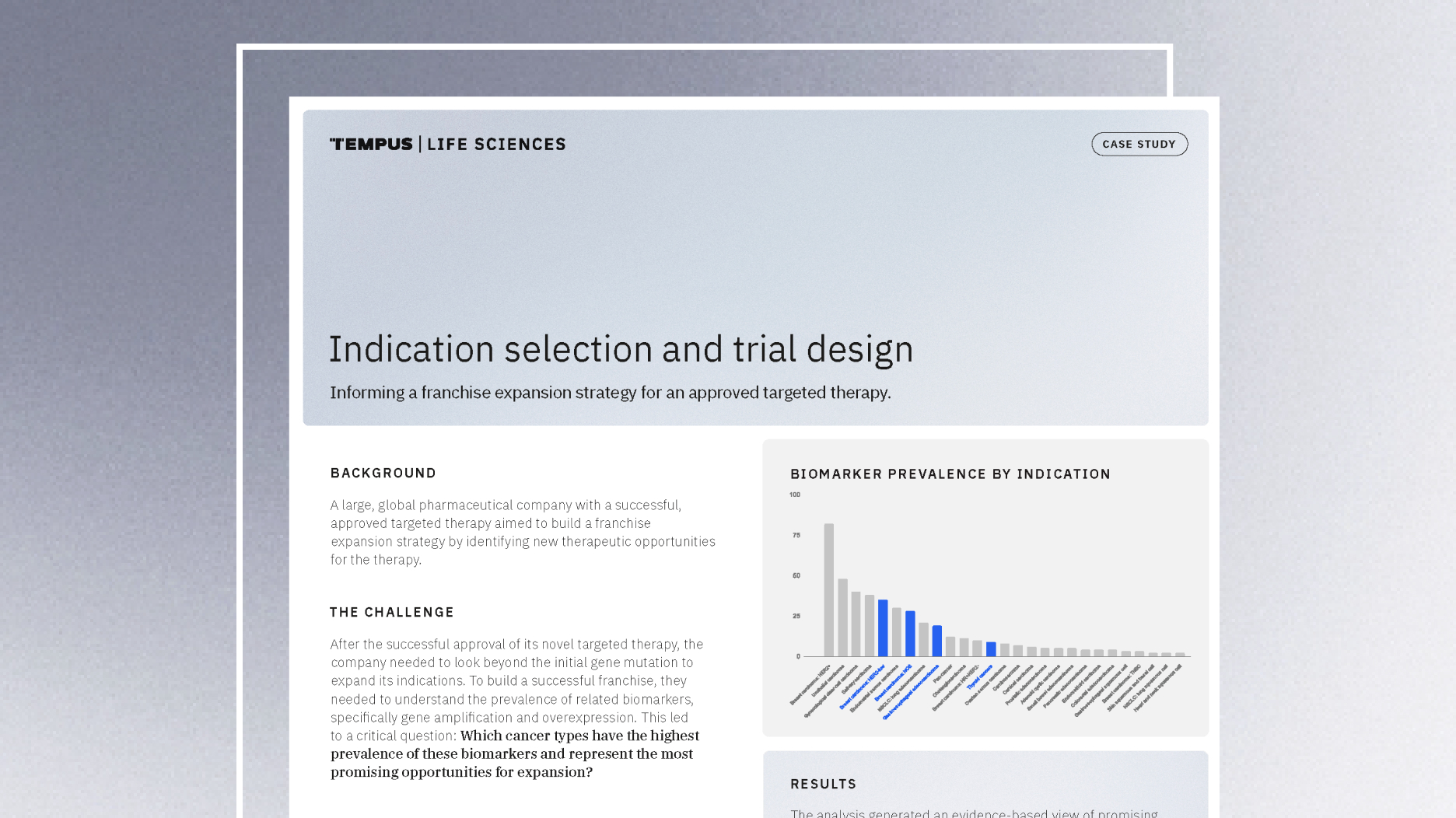
Guiding indication expansion with multimodal real-world data
Discover how a biopharma company used Tempus’ multimodal real-world data to guide its indication expansion strategy. See how our analysis of biomarker prevalence helped them identify new opportunities, prioritize R&D, and inform future trial design.
Read more