-
PROVIDERS
Register now
Are you getting the full picture? A webinar series on the power of comprehensive intelligent diagnostics
-
LIFE SCIENCES
Enroll now
Tempus’ Patient-Derived Organoid ScreensEvaluate the efficacy of your preclinical compounds using fixed organoid panels designed for diverse therapeutic applications. Space is limited — enroll by June 30, 2025, to secure your spot.
-
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
12/01/2023
Radiogenomics For EGFR Mutation Status Prediction in CT Images: Impact Of Model Design on Performance and Prospective Generalizability
RSNA 2023
PRESENTATION
Authors
Jacob Gordon; Omid Haji Maghsoudi; Jovicarole Raya; Babak Rasolzadeh; Martin Stumpe; Marina Codari; Kunal Nagpal
Purpose: We hypothesized that design choices could affect predictive performance and generalizability of radiogenomic models. We investigated the impact of feature extraction and selection approaches on models developed to predict EGFR mutation status (EGFRm+/-) in NSCLC, assessing robustness on two different input CT image types.
Materials and Methods: We analyzed CT images from CT and PET/CT studies of US patients with NSCLC from TCIA (25% EGFRm+). Two feature extraction techniques were compared: hand-crafted features from segmented lesions and deep features from a pre-trained ResNet applied to a bounding box around the largest tumor. Principal component analysis (PCA) or Least Absolute Shrinkage and Selection Operator (LASSO) was used to select 5 features prior to classification by a support vector machine, with model performances evaluated via 5-fold cross validation AUC. Splits were stratified by EGFRm and disease stage. Prospective
generalizability was evaluated using a temporal split (80:20 based on shifted dates). AUCs were reported as median and interquartile range.
Results: When predicting EGFR status from diagnostic CT images (n=171), hand-crafted features selected either with PCA or LASSO resulted in effective stratification: AUC of 0.80 (0.73 – 0.80) and 0.80 (0.69 – 0.82), respectively, and encouraging prospective generalizability (0.86 for PCA and 0.76 for LASSO). By contrast, deep features showed poor performance using either PCA (0.60 [0.60 – 0.67]) or LASSO (0.46 [0.44 – 0.66]) as well as poor prospective generalizability (0.68 and 0.38, respectively). When utilizing low-fidelity CT from PET/CT studies (n=134), hand-crafted features selected with PCA resulted in an AUC of 0.75 (0.73 – 0.77) vs. 0.66 (0.64 – 0.76) for LASSO. However, LASSO showed better prospective generalizability (0.79 vs 0.76). In this dataset, deep features combined with PCA showed improved predictive power compared with LASSO: AUCs of 0.72 (0.69 – 0.8) and 0.47 (0.46 – 0.67), respectively. With regard to prospective generalizability AUCs were 0.73 and 0.44 for PCA and LASSO, respectively.
Conclusion: Performance and prospective generalizability for predicting EGFR mutation status varied widely across design settings. Tumor-based hand-crafted features filtered by PCA performed better than deep feature-based approaches regardless of image type. The effectiveness of the hand-crafted feature models supports the potential of radiogenomics in early identification of patients likely to harbor EGFR mutations.
Clinical Relevance/Application: Treatment planning in NSCLC depends on EGFRm. Radiogenomic models may help to rapidly identify patients who may harbor EGFRm. However, rigorous analysis of model design is key to successful model development.
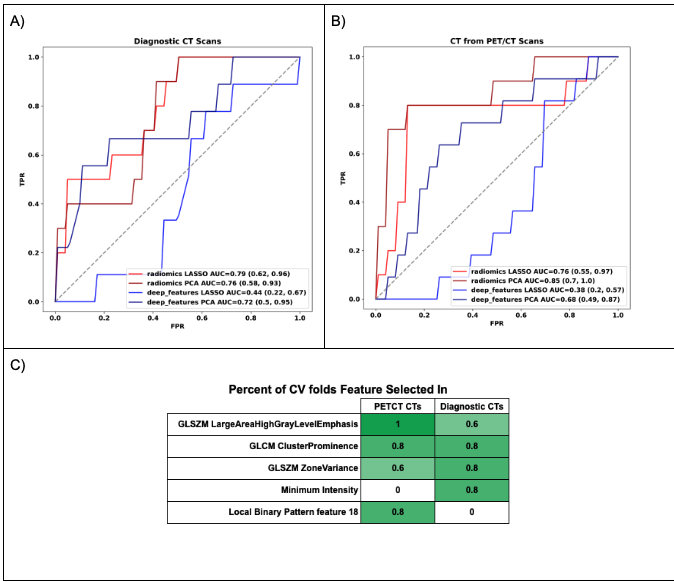
Figure (A-B) Receiver operating characteristic curves for EGFR mutational status prediction on the temporal test set from diagnostic CT scans and staging PET/CTs respectively, comparing different feature extraction (hand-crafted[radiomics] vs. deep feature-based) and selection methods (LASSO vs. PCA). (C) Table comparing the most common hand-crafted radiomics features selected by LASSO for models built with CT images from PET/CT and Diagnostic CT exams respectively.