-
PROVIDERS
New MRD Medicare Coverage for Select Indications*
*When coverage criteria are met. Additional criteria and exceptions for coverage may apply.
-
LIFE SCIENCES
Learn more
Building the engine to scale data abstraction through AI
Learn how Tempus’ AI engine transforms unstructured clinical text into analysis-ready real-world data -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
11/16/2024
Integrated Transcriptomic Analysis of CIC::DUX4 Fusion Sarcomas Reveals Elevated Proliferation Signaling and Implicates DNA Repair Gene Pole
CTOS 2024
PRESENTATION
Authors
Nathan D. Seligson and James L. Chen
Background: CIC::DUX4 fusion sarcomas (CDS) are a distinct subset of high-grade undifferentiated round cell sarcomas characterized by a specific oncogenic translocation involving the CIC and DUX4 genes. These sarcomas are rare and aggressive without effective targeted therapies. Beyond the presence of the disease-defining fusion event, there is limited knowledge regarding the molecular mechanisms driving these sarcomas. In this study, we integrate heretofore unavailable RNA expression data from clinical samples and in vitro models of CDS to identify molecular drivers of this aggressive disease.
Methods: De-identified transcriptomic profiling data of specimens from patients with CDS were collected from the Tempus AI database. Additional data were collected from publically available datasets including GSE60740 (14 CDS), GSE34620 (117 Ewing sarcoma, EwS), and GSE60740 (CDS cell line +/- shRNA CIC::DUX4 knockdown) as previously described. To identify genesets dysregulated by the CIC::DUX4 fusion three comparisons were performed. First, we evaluated over or under expressed pathways within CDS (p<0.05). Next, we evaluated pathway expression between CDS and EwS (p<0.1) and between CDS cell lines with and without shRNA knockdown of CIC::DUX4 (p<0.1), Single sample gene set analysis was performed using the GSVA package in R and both Hallmarks of Cancer and Reactome genesets. Single and two sample T tests were used as appropriate. Humanbase was used for gene interaction analysis.
Results: In total, 21 samples from patients with CDS were evaluable for analysis. Using the Hallmarks of cancer set, our integrated analysis identified seven significantly upregulated molecular pathways, including MYC Targets, G2M Checkpoint, Mitotic Spindle, E2F Targets, and Unfolded Protein Response. This finding implicates cellular proliferation as highly upregulated by the CIC::DUX4 fusion even in comparison to highly proliferative EwS tumors. To identify the genes responsible for driving these significant molecular pathways, we selected the 20 highest expressed genes (intra-tumor z-score >2) in the Tempus CDS clinical samples as potential drivers/targets specific to the CIC::DUX4 fusion. A majority of these genes (15/20, 75%) were associated with replication machinery (e.g. genes coding for ribosomal proteins). Of the remaining genes, only ETV4 and POLE were specific to the CIC::DUX4 fusion across patient samples (CDS vs EwS) and in vitro CDS models (Figure 1A, 1B). While ETV4 is a known driver of CDS, our finding of POLE upregulation is unique and implicates a role of efficient DNA repair in this disease. Using Reactome gensets, we found that the CIC::DUX4 fusion was associated with increased DNA repair activity of all DNA repair pathways tested (Mismatch Repair, Nucleotide Excision Repair, Base Excision Repair, Homologous Recombination, Non-homologous End Joining) in comparison to EwS patient samples and in in vitro CDS models.
Conclusions: Our integrated transcriptomic analysis reveals CDS as highly proliferative and that the CIC::DUX4 fusion drives proliferation through ETV4. Our findings also suggest that an upregulation of DNA repair, possibly through POLE, may be necessary to stabilize these cells through replication stress. Additional studies will be needed for clinical translation in this rare and aggressive malignancy.
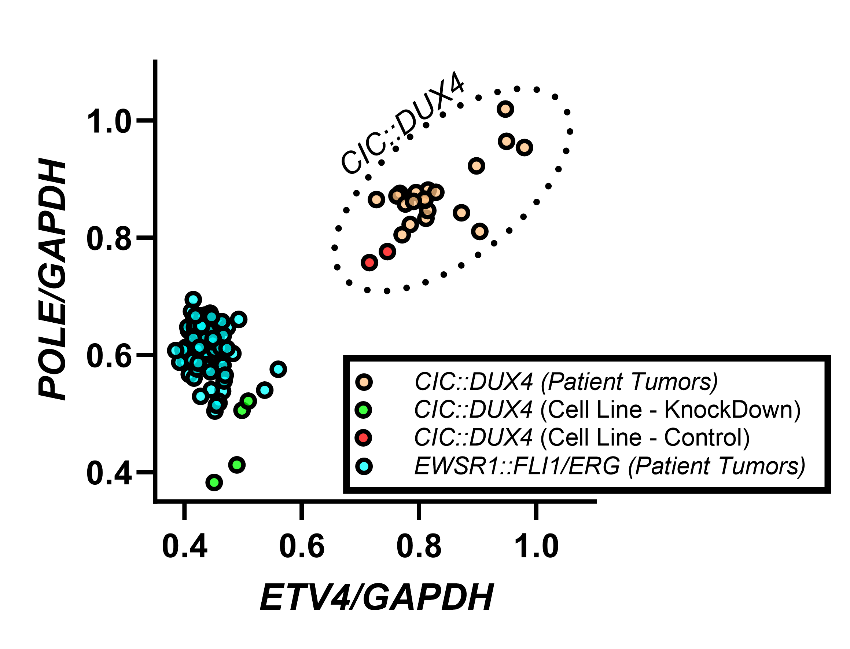
ETV4 and POLE expression are specific to the CIC::DUX4 fusion across patient samples and in vitro models.