-
PROVIDERS
Register now
Are you getting the full picture? A webinar series on the power of comprehensive intelligent diagnostics
-
LIFE SCIENCES
Enroll now
Tempus’ Patient-Derived Organoid ScreensEvaluate the efficacy of your preclinical compounds using fixed organoid panels designed for diverse therapeutic applications. Space is limited — enroll by June 30, 2025, to secure your spot.
-
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
11/05/2024
Self-Supervised Representation Learning Enables Genomic Prediction at Single-Organoid Resolution
SITC 2024
PRESENTATION
Authors
Geoffrey Schau, Stanislaw Sydlo, Brandon Mapes, Elle Moore, Timothy Don Lopez, Michael Streit, Justin Guinney, Sonal Khare, Madhavi Kannan, Chi-Sing Ho
Background Tumor organoids (TO) have emerged as compelling cancer models due in part to their conserved properties of tumor heterogeneity and 3D structure of the tumor-immune microenvironment. A principal challenge in high-throughput screening experiments is tracking and labeling the somatic states of TOs during clonal and subclonal selection and expansion in the presence of immune effector cells. In this work, we train and evaluate multiple computer-vision based models for the identification of driver mutations at single-organoid resolution.
Methods DNA from TO lines was sequenced using the Tempus xT assay. Lines were cultured in the presence of natural killer effector cells and regularly imaged via brightfield confocal microscopy. Individual TOs were segmented from the original image and processed with three feature extractors: an unsupervised pre-trained convolutional neural network (CNN), a self-supervised neural network, and CellProfiler,an industry-leading microscopy analysis software suite. Secondary predictor models were trained on each feature set to predict the mutational status of several cancer driver genes using soft-labels derived from sample-level sequencing. For each secondary predictor, splitting observations into training and testing datasets was stratified by sample, mutation status, and cancer type. Classification performance was evaluated across each of the three feature sets and combinations thereof for each of the intended gene targets. The overall workflow is shown in figure 1.
Results We find that models using human-interpretable features generated by CellProfiler generally outperform both pre-trained CNNs and self-supervised computer vision models in predicting mutation status in TOs, and that combining feature sets generally confers an additive benefit for predicting gene mutation (figure 2). Of the eight genes evaluated in this study, the most performant model was observed in predicting KRAS (AUROC=0.82) and PTEN (AUROC=0.79) and least effective for TP53 (AUROC=0.57). Finally, ranked feature importance scoring suggests that morphological features most useful in predicting mutation status were tumor size, perimeter, and metrics of spatial intensity distribution.
Conclusions We present a single-organoid featurization and classification strategy for single-organoid gene mutation prediction from brightfield morphological features, enabling a high-throughput, low-cost approach for inferring an individual organoids’ somatic state. This approach can be used for rapid assessment of heterogeneous clonality in progenitor cell populations in the context of high-throughput screenings.
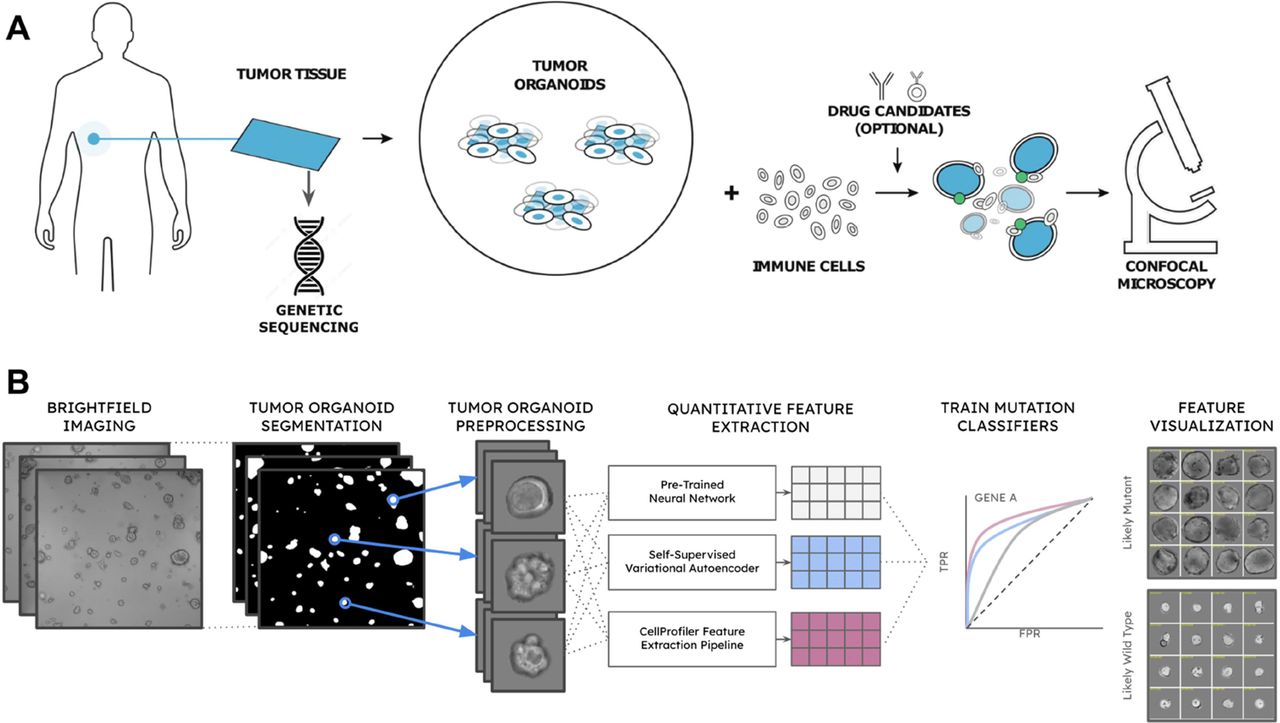
Figure 1. – (A) Patient-derived tumor organoids are grown in the presence of immune cells and optional drug candidates and imaged via time-series confocal microscopy to assess growth kinetics. (B) Analytical pipeline consists of: brightfield image capture; tumor organoid segmentation; organoid bounding box cropping and background subtraction; feature extraction with a pre-trained neural network, a self-supervised neural network, and CellProfiler; and mutational prediction with each extracted feature set; and tumor organoid feature visualization demonstrating individual tumor organoids rank-ordered by the model’s prediction score
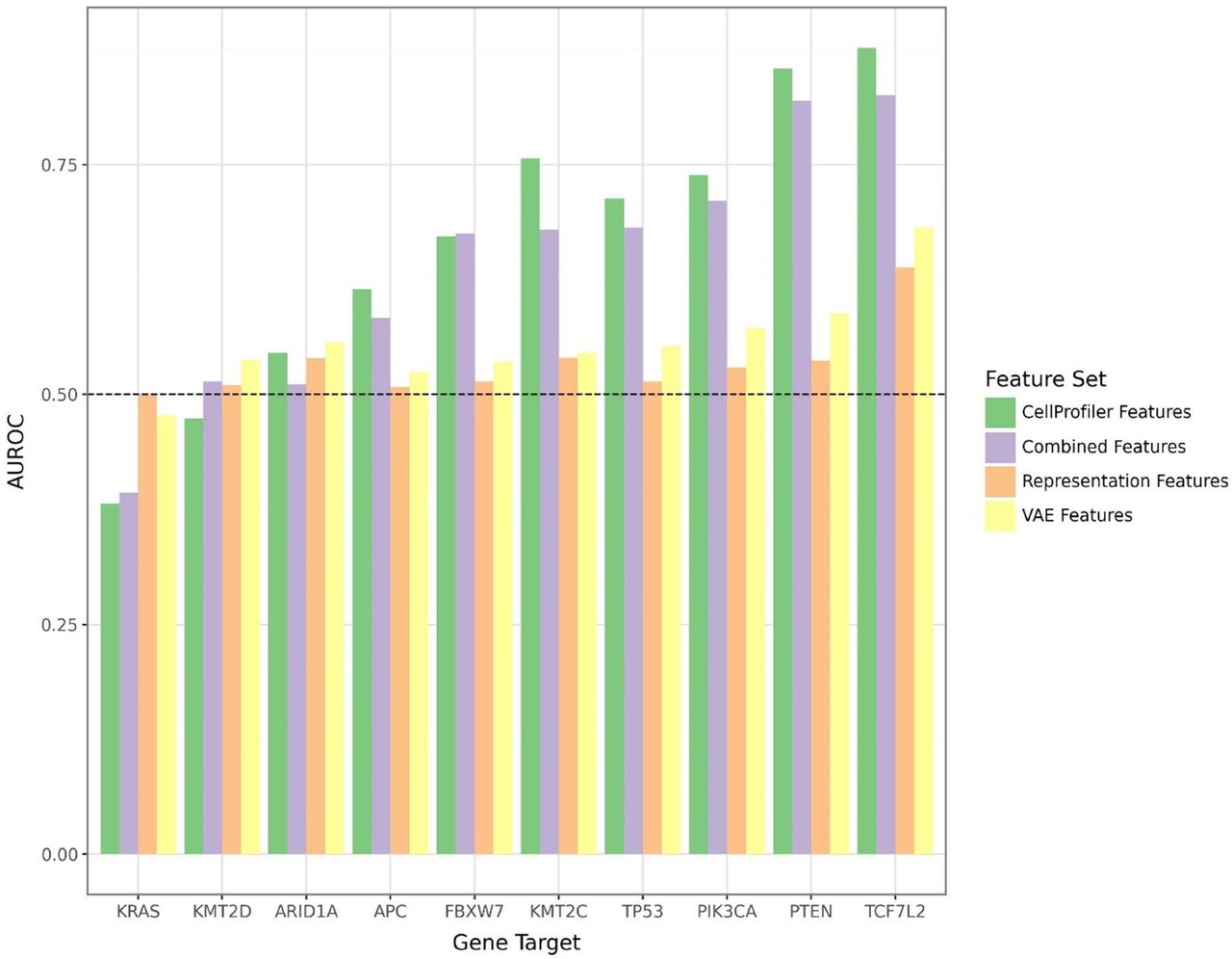
Figure 2 – Area under the receiver operator characteristics (AUROC) curve illustrates differential predictability of gene mutation status with different representation methods within tumor organoids co-cultured with immune cells, demonstrating a trend of marked utility of human-interpretable features generated by CellProfiler relative to unsupervised and self-supervised learning systems