-
PROVIDERS
New MRD Medicare Coverage for Select Indications*
*When coverage criteria are met. Additional criteria and exceptions for coverage may apply.
-
LIFE SCIENCES
Register now
UPCOMING WEBINARTranslating data into an actionable R&D strategy
-
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
11/22/2022
Dual testing in metastatic breast cancer
Tempus research exploring the concordance of pathogenic variants across a variety of criteria for metastatic breast cancer patients.
Authors
Matt MacKay, PhD
Director, Molecular Analytics, Tempus

Director, Molecular Analytics, Tempus

Research summary
Tempus offers both a tissue based NGS assay (Tempus xT) as well as a cell-free DNA (cfDNA) NGS assay (Tempus xF), allowing clinicians flexibility in their diagnostic decisions. However, while each modality offers different benefits, concordance between the two types of tests is a source of ongoing study. To address this, Tempus researchers evaluated the concordance between Tempus xT and Tempus xF relative to cfDNA sampling frequency in a large, clinically annotated deidentified metastatic breast cancer (mBC) data set using the Tempus Lens platform. The team investigated the concordance of pathogenic variants across genes, time between tests, the variant allele frequencies, and mBC subtypes.
The study reviewed the records of 300 patients with stage IV mBC, 94% of which had at least one pathogenic alteration found between ctDNA biopsy and tissue NGS. When tissue and blood collection were ≤ 7 days apart (n=56), 77.8% of pathogenic tissue variants were found within each patient’s cfDNA, and 75.7% of pathogenic cfDNA variants were found within each patient’s tissue, showing high concordance of variant detection for each patient. At the individual variant level, 20% of pathogenic variants were uniquely detected by xF liquid biopsy, while 18% were detected solely in tissue by xT (fig.1). Notably, all ESR1 variants were more likely to be identified when analyzing cfDNA (cfDNA-only or concurrent) than tissue samples alone (fig. 2). Combined testing with xT and xF may provide a more complete picture of the patient’s genetic landscape than either testing method alone.
The concordance observed (62% concordance, ≤7 days) suggest that cfDNA NGS may reliably represent tissue genomics, but the increase in discordance over time also suggests a possible evolution of tumor genomics that can be measured by sequential testing with Tempus xF. Additionally, the detection of unique variants by each testing modality may indicate that using both tests could increase the overall variants detected compared to using either individually.
Next steps
Contact Tempus to discuss the findings with our team, and learn more about concurrent testing in breast cancer patients.
Unique variants may be detected between tissue samples and cfDNA
Figure 1
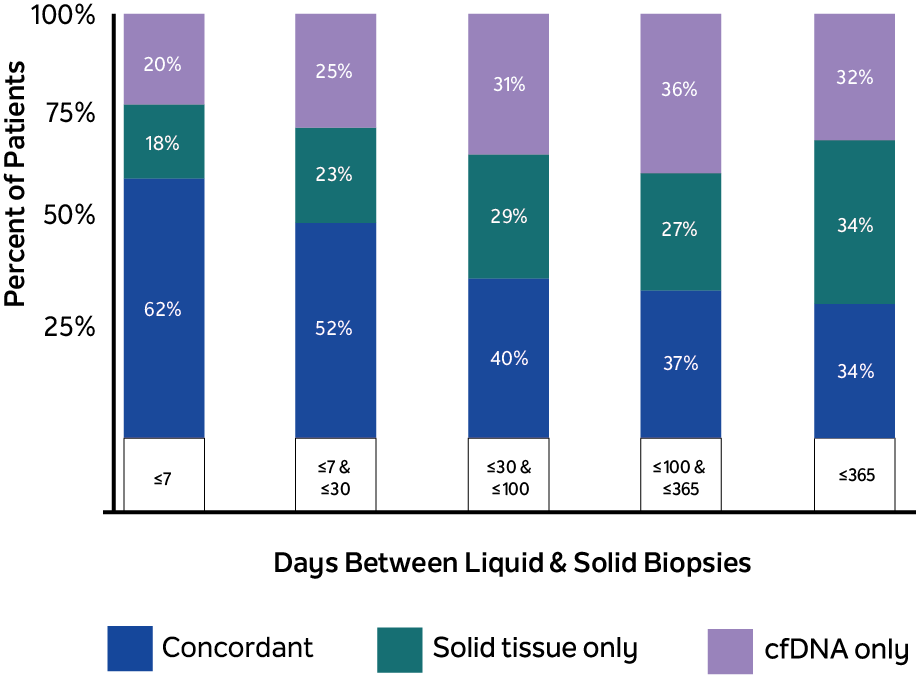
20% of pathogenic variants were uniquely detected by xF liquid biopsy
18% were detected solely in tissue by xT
18% were detected solely in tissue by xT
ESR1 variants were more likely to be identified when analyzing cfDNA (cfDNA-only or concurrent) than tissue samples alone
Figure 2
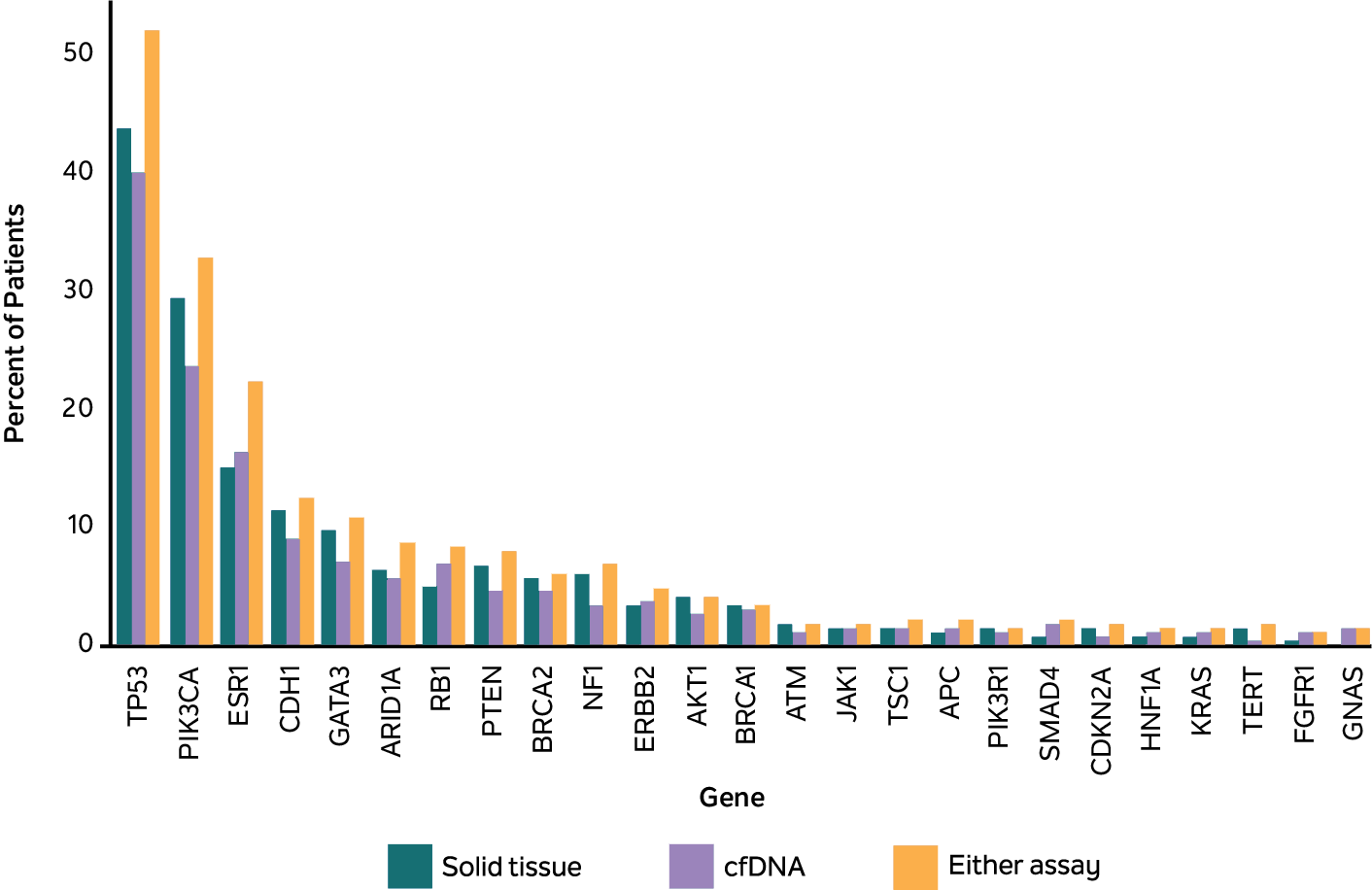
Shown are the % of MBC patients with ≥1 of the top 25 most common pathogenic variants in cfDNA (green), tissue (blue), and either test (purple), as assessed using the Tempus xF, xT, or xF/xT assay and the Tempus LENS platform.
Liu MC, et al. JCO Precis Oncol. 2022;6(1):e2100321.
READ THE MANUSCRIPT
-
02/12/2026
Beyond the EHR: Building a reliable mortality endpoint for oncology research
Emilie Scherrer, Senior Director and Head of Outcomes Research, explains how Tempus’ validated, composite mortality endpoint overcomes data fragmentation to provide a trustworthy foundation for real-world evidence.
Read more -
02/05/2026
Accelerating a phase 1 oncology trial: The TIME Network’s impact on patient enrollment
For life sciences companies, speed and precision are paramount. The ability to efficiently launch a Phase 1 trial and generate early clinical data is fundamental to long-term success. The Tempus TIME Trial Network provides the infrastructure to help achieve these goals.
Read more -
02/05/2026
Building the engine to scale data abstraction through AI
Learn how Tempus’ AI engine, built on agentic architectures, transforms unstructured clinical text into analysis-ready real-world data. Tempus AI leaders detail the multi-layered system that ensures speed, scale, and fidelity.
Read more


